 |
Tina M. Henkin
henkin.3@osu.edu B.A., Biology, Swarthmore College |
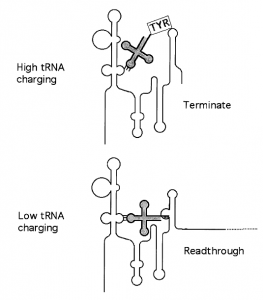
| Department Chair Professor of Microbiology Robert W. and Estelle S. Bingham Professor of Biological Sciences Member, Center for RNA Biology Member, MCDB Member, OSBPRiboswitch RNAs; Transcription termination; Translation initiation; RNA structure/function; antibiotic design; Gram-positive bacteria The main area of interest in our laboratory is the analysis of the mechanisms through which cells sense changes in their environment and transmit that information to the level of gene expression. We use the Gram-positive bacterium Bacillus subtilis as a model system, and we focus primarily on genes involved in protein synthesis and amino acid metabolism. We have uncovered systems in which nascent RNA transcripts act as riboswitches to directly sense physiological signals and control gene expression through RNA structural rearrangements. Nascent RNAs can sense uncharged tRNA: the T box system Characterization of the B. subtilis tyrS gene, encoding tyrosyl-tRNA synthetase, revealed a novel mechanism of gene regulation at the level of transcription antitermination. The tyrS gene is a member of a large family of aminoacyl-tRNA synthetase and amino acid biosynthesis genes in Gram-positive bacteria that are regulated by a common mechanism. Each gene in this family responds individually to limitation for the appropriate amino acid. Amino acid limitation is monitored via interaction of the 5’ region of the nascent transcript with the cognate uncharged tRNA. This interaction is directed by pairing of the anticodon of the tRNA with a single codon, designated the “Specifier Sequence,” in the mRNA. The mRNA-tRNA interaction occurs in the absence of translation, and antitermination can occur in a purified transcription system with no additional cellular factors, indicating that the mRNA is sufficient for specific recognition of the cognate tRNA. We are currently investigating the molecular details of the leader RNA-tRNA interaction, and the structural shifts in both RNA partners that occur upon binding. We are also testing novel antibiotics for their ability to target the T box mechanism.
Nascent RNAs can sense small molecules: metabolite-binding riboswitch RNAs Analysis of genes involved in methionine metabolism revealed a second global transcription antitermination system, dedicated to genes in this pathway. Like the T box system, the S box system is widely used in Gram-positive organisms. Genes regulated by this mechanism contain highly conserved sequence and structural elements in their mRNAs, and expression is induced by starvation for methionine. We have now shown that the molecular effector for this system is S -adenosylmethionine, which binds directly to the leader RNA and modululates its structure to promote transcription termination. A second SAM-binding RNA, the SMK box, was identified in lactic acid bacteria and shown to regulate gene expression at the level of translation initiation. We have also shown that lysine biosynthesis genes are regulated by a similar mechanism, with specific leader RNA binding of lysine. Current work is focusing on the molecular mechanisms of effector recognition and RNA rearrangement in response to effector binding. |
|


