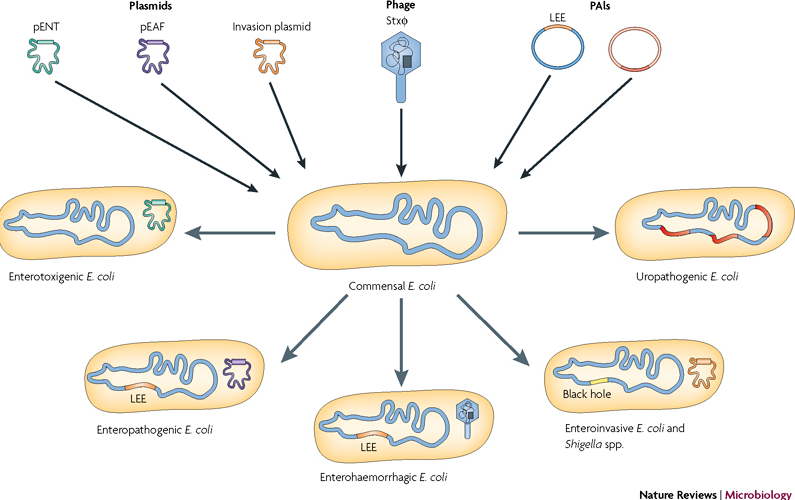
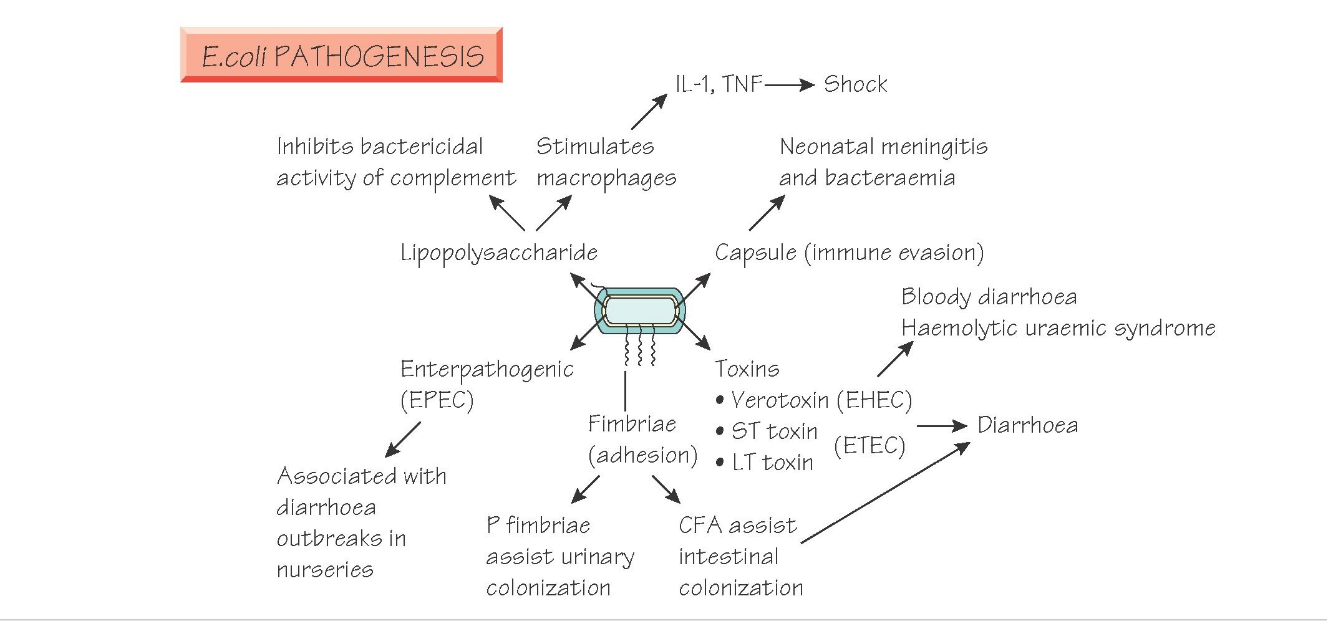
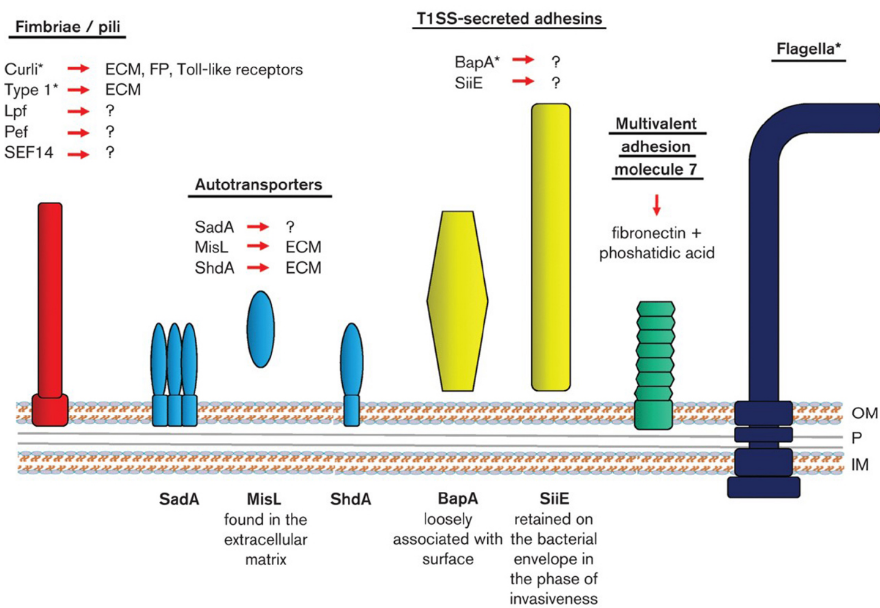
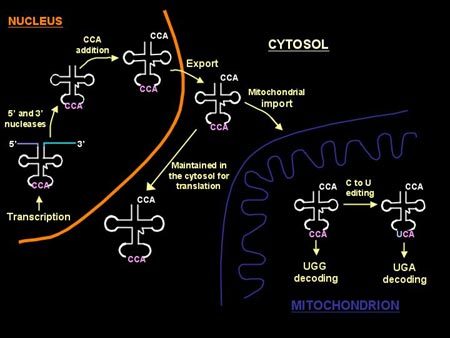
Bacteriophage Ecology and Evolutionary Ecology
Stephen T. Abedon abedon.1@osu.edu Associate Professor. Ph.D., University of Arizona, 1990. Bacteriophage Ecology and Evolutionary Ecology Bacteriophage, or phage, are the viruses of bacteria. I am interested especially in phage adaptation: How phages can and have optimized their characteristics toward meeting the challenges of growth and survival. My research