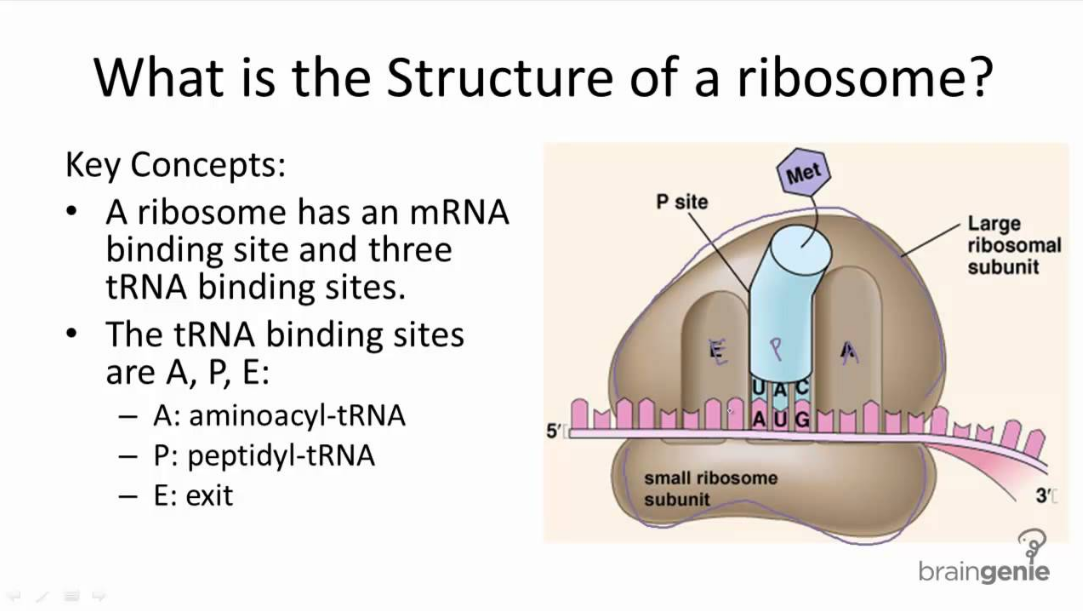
Understanding the Relationship Between Ribosome Structure and Function
Kurt Fredrick fredrick.5@osu.edu Assistant Professor B.A. Biology, Gustavus Adolphus College, 1992 Ph.D. Microbiology, Cornell University, 1997 Postdoctoral research, University of California Santa Cruz, 1997-2003 We are interested in how the ribosome works. The ribosome is a large (~2.5 MDa), two-subunit, RNA-based machine that translates the genetic code in